Code
import numpy as np
# declare a vector using a list as the argument
v = np.array([1, 2.0, 3, 4])
varray([1., 2., 3., 4.])numpyNumPy is the fundamental package for scientific computing with Python. It contains among other things:
Besides its obvious scientific uses, NumPy can also be used as an efficient multi-dimensional container for general data. Arbitrary data-types can be defined. This allows NumPy to seamlessly and speedily integrate with a wide variety of databases.
Library documentation: http://numpy.org/
numpy.array objectimport numpy as np
# declare a vector using a list as the argument
v = np.array([1, 2.0, 3, 4])
varray([1., 2., 3., 4.])list([1, 2.0, 3, 4])[1, 2.0, 3, 4]type(v)numpy.ndarrayv.shape(4,)v.ndim1v.dtype is floatFalsev.dtype dtype('float64')np.uint8 is intFalseUse copilot explain to understand the chunks:
The np.uint8 is a data type in NumPy, representing an unsigned 8-bit integer, which can store values from 0 to 255. The int type is the built-in integer type in Python, which can represent any integer value without a fixed size limit.
np.array([2**120, 2**40], dtype=np.int64)--------------------------------------------------------------------------- OverflowError Traceback (most recent call last) Cell In[9], line 1 ----> 1 np.array([2**120, 2**40], dtype=np.int64) OverflowError: Python int too large to convert to C long
np.uint16 is int Falsenp.uint32 is intFalsew = np.array([1.3, 2, 3, 4], dtype=np.int64)
warray([1, 2, 3, 4])w.dtypedtype('int64')a = np.arange(100)type(a)numpy.ndarraynp.array(range(100))array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84,
85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99])aarray([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84,
85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99])a.dtypedtype('int64')-3 * a ** 2array([ 0, -3, -12, -27, -48, -75, -108, -147,
-192, -243, -300, -363, -432, -507, -588, -675,
-768, -867, -972, -1083, -1200, -1323, -1452, -1587,
-1728, -1875, -2028, -2187, -2352, -2523, -2700, -2883,
-3072, -3267, -3468, -3675, -3888, -4107, -4332, -4563,
-4800, -5043, -5292, -5547, -5808, -6075, -6348, -6627,
-6912, -7203, -7500, -7803, -8112, -8427, -8748, -9075,
-9408, -9747, -10092, -10443, -10800, -11163, -11532, -11907,
-12288, -12675, -13068, -13467, -13872, -14283, -14700, -15123,
-15552, -15987, -16428, -16875, -17328, -17787, -18252, -18723,
-19200, -19683, -20172, -20667, -21168, -21675, -22188, -22707,
-23232, -23763, -24300, -24843, -25392, -25947, -26508, -27075,
-27648, -28227, -28812, -29403])a[42] = 13a[42] = 1025np.info(np.int16) int16()
Signed integer type, compatible with C ``short``.
:Character code: ``'h'``
:Canonical name: `numpy.short`
:Alias on this platform (Linux x86_64): `numpy.int16`: 16-bit signed integer (``-32_768`` to ``32_767``).
Methods:
all -- Scalar method identical to the corresponding array attribute.
any -- Scalar method identical to the corresponding array attribute.
argmax -- Scalar method identical to the corresponding array attribute.
argmin -- Scalar method identical to the corresponding array attribute.
argsort -- Scalar method identical to the corresponding array attribute.
astype -- Scalar method identical to the corresponding array attribute.
bit_count -- int16.bit_count() -> int
byteswap -- Scalar method identical to the corresponding array attribute.
choose -- Scalar method identical to the corresponding array attribute.
clip -- Scalar method identical to the corresponding array attribute.
compress -- Scalar method identical to the corresponding array attribute.
conj -- None
conjugate -- Scalar method identical to the corresponding array attribute.
copy -- Scalar method identical to the corresponding array attribute.
cumprod -- Scalar method identical to the corresponding array attribute.
cumsum -- Scalar method identical to the corresponding array attribute.
diagonal -- Scalar method identical to the corresponding array attribute.
dump -- Scalar method identical to the corresponding array attribute.
dumps -- Scalar method identical to the corresponding array attribute.
fill -- Scalar method identical to the corresponding array attribute.
flatten -- Scalar method identical to the corresponding array attribute.
getfield -- Scalar method identical to the corresponding array attribute.
is_integer -- integer.is_integer() -> bool
item -- Scalar method identical to the corresponding array attribute.
max -- Scalar method identical to the corresponding array attribute.
mean -- Scalar method identical to the corresponding array attribute.
min -- Scalar method identical to the corresponding array attribute.
nonzero -- Scalar method identical to the corresponding array attribute.
prod -- Scalar method identical to the corresponding array attribute.
put -- Scalar method identical to the corresponding array attribute.
ravel -- Scalar method identical to the corresponding array attribute.
repeat -- Scalar method identical to the corresponding array attribute.
reshape -- Scalar method identical to the corresponding array attribute.
resize -- Scalar method identical to the corresponding array attribute.
round -- Scalar method identical to the corresponding array attribute.
searchsorted -- Scalar method identical to the corresponding array attribute.
setfield -- Scalar method identical to the corresponding array attribute.
setflags -- Scalar method identical to the corresponding array attribute.
sort -- Scalar method identical to the corresponding array attribute.
squeeze -- Scalar method identical to the corresponding array attribute.
std -- Scalar method identical to the corresponding array attribute.
sum -- Scalar method identical to the corresponding array attribute.
swapaxes -- Scalar method identical to the corresponding array attribute.
take -- Scalar method identical to the corresponding array attribute.
to_device -- None
tobytes -- None
tofile -- Scalar method identical to the corresponding array attribute.
tolist -- Scalar method identical to the corresponding array attribute.
tostring -- Scalar method identical to the corresponding array attribute.
trace -- Scalar method identical to the corresponding array attribute.
transpose -- Scalar method identical to the corresponding array attribute.
var -- Scalar method identical to the corresponding array attribute.
view -- Scalar method identical to the corresponding array attribute.np.int16numpy.int16dict(enumerate(a)){0: np.int64(0),
1: np.int64(1),
2: np.int64(2),
3: np.int64(3),
4: np.int64(4),
5: np.int64(5),
6: np.int64(6),
7: np.int64(7),
8: np.int64(8),
9: np.int64(9),
10: np.int64(10),
11: np.int64(11),
12: np.int64(12),
13: np.int64(13),
14: np.int64(14),
15: np.int64(15),
16: np.int64(16),
17: np.int64(17),
18: np.int64(18),
19: np.int64(19),
20: np.int64(20),
21: np.int64(21),
22: np.int64(22),
23: np.int64(23),
24: np.int64(24),
25: np.int64(25),
26: np.int64(26),
27: np.int64(27),
28: np.int64(28),
29: np.int64(29),
30: np.int64(30),
31: np.int64(31),
32: np.int64(32),
33: np.int64(33),
34: np.int64(34),
35: np.int64(35),
36: np.int64(36),
37: np.int64(37),
38: np.int64(38),
39: np.int64(39),
40: np.int64(40),
41: np.int64(41),
42: np.int64(1025),
43: np.int64(43),
44: np.int64(44),
45: np.int64(45),
46: np.int64(46),
47: np.int64(47),
48: np.int64(48),
49: np.int64(49),
50: np.int64(50),
51: np.int64(51),
52: np.int64(52),
53: np.int64(53),
54: np.int64(54),
55: np.int64(55),
56: np.int64(56),
57: np.int64(57),
58: np.int64(58),
59: np.int64(59),
60: np.int64(60),
61: np.int64(61),
62: np.int64(62),
63: np.int64(63),
64: np.int64(64),
65: np.int64(65),
66: np.int64(66),
67: np.int64(67),
68: np.int64(68),
69: np.int64(69),
70: np.int64(70),
71: np.int64(71),
72: np.int64(72),
73: np.int64(73),
74: np.int64(74),
75: np.int64(75),
76: np.int64(76),
77: np.int64(77),
78: np.int64(78),
79: np.int64(79),
80: np.int64(80),
81: np.int64(81),
82: np.int64(82),
83: np.int64(83),
84: np.int64(84),
85: np.int64(85),
86: np.int64(86),
87: np.int64(87),
88: np.int64(88),
89: np.int64(89),
90: np.int64(90),
91: np.int64(91),
92: np.int64(92),
93: np.int64(93),
94: np.int64(94),
95: np.int64(95),
96: np.int64(96),
97: np.int64(97),
98: np.int64(98),
99: np.int64(99)}a + 1array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 1026, 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99,
100])b = a + 1
barray([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 1026, 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99,
100])a is bFalsef = id(a)
a += 1
f, id(a)(127545041510032, 127545041510032)aarray([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 1026, 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99,
100])barray([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 1026, 44,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99,
100])Beware of the dimensions: a 1D array is not the same as a 2D array with 1 column
a1 = np.array([1, 2, 3])
print(a1, a1.shape, a1.ndim)[1 2 3] (3,) 1a2 = np.array([1, 2, 3])
print(a2, a2.shape, a2.ndim)[1 2 3] (3,) 1List the attributes and methods of class numpy.ndarray. You may use function dir() and filter the result using methods for objects of class string.
a2.dot(a1) # inner product np.int64(14)(
np.array([a2])
.transpose() # column vector
.dot(np.array([a1]))
) # column vector multiplied by row vectorarray([[1, 2, 3],
[2, 4, 6],
[3, 6, 9]])(
np.array([a2])
.transpose()#.shape
)array([[1],
[2],
[3]])(
a2.reshape(3,1) # all explicit
.dot(a1.reshape(1, 3))
)array([[1, 2, 3],
[2, 4, 6],
[3, 6, 9]])# Declare a 2D array using a nested list as the constructor argument
M = np.array([[1,2],
[3,4],
[3.14, -9.17]])
Marray([[ 1. , 2. ],
[ 3. , 4. ],
[ 3.14, -9.17]])M.shape, M.size((3, 2), 6)M.ravel(), M.ndim, M.ravel().shape(array([ 1. , 2. , 3. , 4. , 3.14, -9.17]), 2, (6,))# arguments: start, stop, step
x = (
np.arange(12)
.reshape(4, 3)
)
xarray([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11]])y = np.arange(3).reshape(3,1)
yarray([[0],
[1],
[2]])x @ y, x.dot(y)(array([[ 5],
[14],
[23],
[32]]),
array([[ 5],
[14],
[23],
[32]]))np.linspace(0, 10, 51) # meaning of the 3 positional parameters ? array([ 0. , 0.2, 0.4, 0.6, 0.8, 1. , 1.2, 1.4, 1.6, 1.8, 2. ,
2.2, 2.4, 2.6, 2.8, 3. , 3.2, 3.4, 3.6, 3.8, 4. , 4.2,
4.4, 4.6, 4.8, 5. , 5.2, 5.4, 5.6, 5.8, 6. , 6.2, 6.4,
6.6, 6.8, 7. , 7.2, 7.4, 7.6, 7.8, 8. , 8.2, 8.4, 8.6,
8.8, 9. , 9.2, 9.4, 9.6, 9.8, 10. ])np.logspace(0, 10, 11, base=np.e), np.e**(np.arange(11))(array([1.00000000e+00, 2.71828183e+00, 7.38905610e+00, 2.00855369e+01,
5.45981500e+01, 1.48413159e+02, 4.03428793e+02, 1.09663316e+03,
2.98095799e+03, 8.10308393e+03, 2.20264658e+04]),
array([1.00000000e+00, 2.71828183e+00, 7.38905610e+00, 2.00855369e+01,
5.45981500e+01, 1.48413159e+02, 4.03428793e+02, 1.09663316e+03,
2.98095799e+03, 8.10308393e+03, 2.20264658e+04]))import matplotlib.pyplot as plt
# Random standard Gaussian numbers
fig = plt.figure(figsize=(8, 4))
wn = np.random.randn(1000)
bm = wn.cumsum()
plt.plot(bm, lw=3)
np.diag(np.arange(10))array([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 1, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 2, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 3, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 4, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 5, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 6, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 7, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 8, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 9]])zozo = np.zeros((10, 10), dtype=np.float32)
zozoarray([[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 0., 0.]], dtype=float32)zozo.shape(10, 10)print(M)[[ 1. 2. ]
[ 3. 4. ]
[ 3.14 -9.17]]M[1, 1]np.float64(4.0)# assign new value
M[0, 0] = 7
M[:, 0] = 42
Marray([[42. , 2. ],
[42. , 4. ],
[42. , -9.17]])Marray([[42. , 2. ],
[42. , 4. ],
[42. , -9.17]])# Warning: the next m is a **view** on M.
# One again, no copies unless you ask for one!
m = M[0, :]
marray([42., 2.])m[:] = 3.14
Marray([[ 3.14, 3.14],
[42. , 4. ],
[42. , -9.17]])m[:] = 7
Marray([[ 7. , 7. ],
[42. , 4. ],
[42. , -9.17]])# slicing works just like with anything else (lists, etc.)
A = np.array([1, 2, 3, 4, 5])
print(A)
print(A[::-1])
print(A[::2])
print(A[:-1:2])[1 2 3 4 5]
[5 4 3 2 1]
[1 3 5]
[1 3][[n + m * 10 for n in range(5)] for m in range(5)][[0, 1, 2, 3, 4],
[10, 11, 12, 13, 14],
[20, 21, 22, 23, 24],
[30, 31, 32, 33, 34],
[40, 41, 42, 43, 44]]A = np.array([[n + m * 10 for n in range(5)] for m in range(5)])
Aarray([[ 0, 1, 2, 3, 4],
[10, 11, 12, 13, 14],
[20, 21, 22, 23, 24],
[30, 31, 32, 33, 34],
[40, 41, 42, 43, 44]])print(A[1:4])[[10 11 12 13 14]
[20 21 22 23 24]
[30 31 32 33 34]]m = A[:, 1:4]m[1, 1] = 123Aarray([[ 0, 1, 2, 3, 4],
[ 10, 11, 123, 13, 14],
[ 20, 21, 22, 23, 24],
[ 30, 31, 32, 33, 34],
[ 40, 41, 42, 43, 44]])A[1]array([ 10, 11, 123, 13, 14])A[:, 1]array([ 1, 11, 21, 31, 41])A[:, ::-1]array([[ 4, 3, 2, 1, 0],
[ 14, 13, 123, 11, 10],
[ 24, 23, 22, 21, 20],
[ 34, 33, 32, 31, 30],
[ 44, 43, 42, 41, 40]])print(A)[[ 0 1 2 3 4]
[ 10 11 123 13 14]
[ 20 21 22 23 24]
[ 30 31 32 33 34]
[ 40 41 42 43 44]]row_indices = np.array([1, 2, 4])
print(A[row_indices])[[ 10 11 123 13 14]
[ 20 21 22 23 24]
[ 40 41 42 43 44]]A[:, row_indices]array([[ 1, 2, 4],
[ 11, 123, 14],
[ 21, 22, 24],
[ 31, 32, 34],
[ 41, 42, 44]])Another way is through masking with an array of bools
# index masking
B = np.arange(5)
row_mask = np.array([True, False, True, False, False])
print(B)
print(B[row_mask])[0 1 2 3 4]
[0 2]A, A[row_mask] , A[:,row_mask](array([[ 0, 1, 2, 3, 4],
[ 10, 11, 123, 13, 14],
[ 20, 21, 22, 23, 24],
[ 30, 31, 32, 33, 34],
[ 40, 41, 42, 43, 44]]),
array([[ 0, 1, 2, 3, 4],
[20, 21, 22, 23, 24]]),
array([[ 0, 2],
[ 10, 123],
[ 20, 22],
[ 30, 32],
[ 40, 42]]))Don’t forget that python does not make copies unless told to do so (same as with any mutable type)
If you are not careful enough, this typically leads to a lot of errors and to being fired !!
y = x = np.arange(6)
x[2] = 123
yarray([ 0, 1, 123, 3, 4, 5])x is yTrue# A real copy
y = x.copy()
x is y False# Or equivalently (but the one above is better...)
y = np.copy(x)x[0] = -12
print(x, y, x is y)[-12 1 123 3 4 5] [ 0 1 123 3 4 5] FalseTo put values of x in y (copy values into an existing array) use
x = np.random.randn(10)
x, id(x)(array([ 0.32317203, 0.622498 , -0.3263707 , 1.02603136, -0.12407659,
-0.71060304, -0.56917053, -1.30891828, -0.36061646, 0.65732983]),
127545039075344)x.fill(2.78) # in place.
x, id(x)(array([2.78, 2.78, 2.78, 2.78, 2.78, 2.78, 2.78, 2.78, 2.78, 2.78]),
127545039075344)x[:] = 3.14 # x.fill(3.14) can. be chained ...
x, id(x)(array([3.14, 3.14, 3.14, 3.14, 3.14, 3.14, 3.14, 3.14, 3.14, 3.14]),
127545039075344)x[:] = np.random.randn(x.shape[0])
x, id(x)(array([-0.14276168, 0.24659027, -0.59410607, -0.76317396, 1.47435406,
0.12526268, 2.16149477, 1.14258093, -1.68895919, -1.42469239]),
127545039075344)y = np.empty(x.shape) # how does empty() work ?
y, id(y)(array([0.14276168, 0.24659027, 0.59410607, 0.76317396, 1.47435406,
0.12526268, 2.16149477, 1.14258093, 1.68895919, 1.42469239]),
127545039076208)y = x
y, id(y), id(x), y is x(array([-0.14276168, 0.24659027, -0.59410607, -0.76317396, 1.47435406,
0.12526268, 2.16149477, 1.14258093, -1.68895919, -1.42469239]),
127545039075344,
127545039075344,
True)In the next line you copy the values of x into an existing array y (of same size…)
y = np.zeros(x.shape)
y[:] = x
y, y is x, np.all(y==x)(array([-0.14276168, 0.24659027, -0.59410607, -0.76317396, 1.47435406,
0.12526268, 2.16149477, 1.14258093, -1.68895919, -1.42469239]),
False,
np.True_)While in the next line, you are aliasing, you are giving a new name y to the object named x (you should never, ever write something like this)
y = x
y is xTrueA numpy array can contain other things than numeric types
arr = np.array(['Labore', 'neque', 'ipsum', 'ut', 'non', 'quiquia', 'dolore.'])
arr, arr.shape, arr.dtype(array(['Labore', 'neque', 'ipsum', 'ut', 'non', 'quiquia', 'dolore.'],
dtype='<U7'),
(7,),
dtype('<U7'))# arr.sum()"_".join(arr)'Labore_neque_ipsum_ut_non_quiquia_dolore.'arr.dtypedtype('<U7')numpySo far, we have only used array or ndarray objects
The is another type: the matrix type
In words: don’t use it (IMhO) and stick with arrays
# Matrix VS array objects in numpy
m1 = np.matrix(np.arange(3))
m2 = np.matrix(np.arange(3))
m1, m2(matrix([[0, 1, 2]]), matrix([[0, 1, 2]]))m1.transpose() @ m2, m1.shape, m1.transpose() * m2(matrix([[0, 0, 0],
[0, 1, 2],
[0, 2, 4]]),
(1, 3),
matrix([[0, 0, 0],
[0, 1, 2],
[0, 2, 4]]))a1 = np.arange(3)
a2 = np.arange(3)
a1, a2(array([0, 1, 2]), array([0, 1, 2]))m1 * m2.T, m1.dot(m2.T)(matrix([[5]]), matrix([[5]]))a1 * a2array([0, 1, 4])a1.dot(a2)np.int64(5)np.outer(a1, a2)array([[0, 0, 0],
[0, 1, 2],
[0, 2, 4]])from scipy.sparse import csc_matrix, csr_matrix, coo_matrixprobs = np.full(fill_value=1/4, shape=(4,))
probsarray([0.25, 0.25, 0.25, 0.25])X = np.random.multinomial(n=2, pvals=probs, size=4) # check you understand what is going on
Xarray([[1, 0, 1, 0],
[0, 0, 2, 0],
[0, 2, 0, 0],
[0, 0, 2, 0]])probsarray([0.25, 0.25, 0.25, 0.25])X_coo = coo_matrix(X) ## coordinate formatprint(X_coo)
X_coo<COOrdinate sparse matrix of dtype 'int64'
with 5 stored elements and shape (4, 4)>
Coords Values
(0, 0) 1
(0, 2) 1
(1, 2) 2
(2, 1) 2
(3, 2) 2<COOrdinate sparse matrix of dtype 'int64'
with 5 stored elements and shape (4, 4)>X_coo.nnz # number pf non-zero coordinates 5print(X, end='\n----\n')
print(X_coo.data, end='\n----\n')
print(X_coo.row, end='\n----\n')
print(X_coo.col, end='\n----\n')[[1 0 1 0]
[0 0 2 0]
[0 2 0 0]
[0 0 2 0]]
----
[1 1 2 2 2]
----
[0 0 1 2 3]
----
[0 2 2 1 2]
----There is also
csr_matrix: sparse rows formatcsc_matrix: sparse columns formatSparse rows is often used for machine learning: sparse features vectors
But sparse column format useful as well (e.g. coordinate gradient descent)
X = np.random.randn(5, 5)
Xarray([[-0.27934931, 0.90292038, 0.18346534, 0.32393199, -0.52303084],
[ 0.06582062, 0.93522102, -0.1966154 , 1.89469087, 1.84690246],
[ 0.77167122, -0.43543013, -0.80626988, -2.41875204, 1.80564412],
[-0.08412047, 1.13484189, 1.17345899, -1.65737546, 0.48533065],
[ 1.01824172, 1.56244786, 1.13970768, 1.12889388, 0.67800411]])# All number displayed by numpy (in the current kernel) are with 3 decimals max
np.set_printoptions(precision=3)
print(X)
np.set_printoptions(precision=8)[[-0.279 0.903 0.183 0.324 -0.523]
[ 0.066 0.935 -0.197 1.895 1.847]
[ 0.772 -0.435 -0.806 -2.419 1.806]
[-0.084 1.135 1.173 -1.657 0.485]
[ 1.018 1.562 1.14 1.129 0.678]]numpy arrays can have any number of dimension (hence the name ndarray)
X = np.arange(18).reshape(3, 2, 3)
Xarray([[[ 0, 1, 2],
[ 3, 4, 5]],
[[ 6, 7, 8],
[ 9, 10, 11]],
[[12, 13, 14],
[15, 16, 17]]])X.shape(3, 2, 3)X.ndim3Visit https://numpy.org/doc/stable/reference/arrays.ndarray.html#arrays-ndarray
A = np.arange(42).reshape(7, 6)
Aarray([[ 0, 1, 2, 3, 4, 5],
[ 6, 7, 8, 9, 10, 11],
[12, 13, 14, 15, 16, 17],
[18, 19, 20, 21, 22, 23],
[24, 25, 26, 27, 28, 29],
[30, 31, 32, 33, 34, 35],
[36, 37, 38, 39, 40, 41]])A.sum(), 42 * 41 //2(np.int64(861), 861)A[:, 3].mean(), np.mean (3 + np.arange(0, 42, 6))(np.float64(21.0), np.float64(21.0))A.mean(axis=0)array([18., 19., 20., 21., 22., 23.])A.mean(axis=1)array([ 2.5, 8.5, 14.5, 20.5, 26.5, 32.5, 38.5])A[:,3].std(), A[:,3].var()(np.float64(12.0), np.float64(144.0))A[:,3].min(), A[:,3].max()(np.int64(3), np.int64(39))A.cumsum(axis=0)array([[ 0, 1, 2, 3, 4, 5],
[ 6, 8, 10, 12, 14, 16],
[ 18, 21, 24, 27, 30, 33],
[ 36, 40, 44, 48, 52, 56],
[ 60, 65, 70, 75, 80, 85],
[ 90, 96, 102, 108, 114, 120],
[126, 133, 140, 147, 154, 161]])Aarray([[ 0, 1, 2, 3, 4, 5],
[ 6, 7, 8, 9, 10, 11],
[12, 13, 14, 15, 16, 17],
[18, 19, 20, 21, 22, 23],
[24, 25, 26, 27, 28, 29],
[30, 31, 32, 33, 34, 35],
[36, 37, 38, 39, 40, 41]])# sum of diagonal
A.trace()np.int64(105)A = np.arange(30).reshape(6, 5)
v1 = np.arange(0, 5)
v2 = np.arange(5, 10)Aarray([[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19],
[20, 21, 22, 23, 24],
[25, 26, 27, 28, 29]])v1, v2(array([0, 1, 2, 3, 4]), array([5, 6, 7, 8, 9]))v1 * v2array([ 0, 6, 14, 24, 36])v1.dot(v2), np.sum(v1* v2)(np.int64(80), np.int64(80))v1.reshape(5,1) @ v2.reshape(1,5)array([[ 0, 0, 0, 0, 0],
[ 5, 6, 7, 8, 9],
[10, 12, 14, 16, 18],
[15, 18, 21, 24, 27],
[20, 24, 28, 32, 36]])# Inner product between vectors
print(v1.dot(v2))
# You can use also (but first solution is better)
print(np.dot(v1, v2))80
80A, v1(array([[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19],
[20, 21, 22, 23, 24],
[25, 26, 27, 28, 29]]),
array([0, 1, 2, 3, 4]))A.shape, v1.shape((6, 5), (5,))# Matrix-vector inner product
A.dot(v1)array([ 30, 80, 130, 180, 230, 280])# Transpose
A.Tarray([[ 0, 5, 10, 15, 20, 25],
[ 1, 6, 11, 16, 21, 26],
[ 2, 7, 12, 17, 22, 27],
[ 3, 8, 13, 18, 23, 28],
[ 4, 9, 14, 19, 24, 29]])print(v1)
# Inline operations (same for *=, /=, -=)
v1 += 2[0 1 2 3 4]A = np.array([[42,2,3], [4,5,6], [7,8,9]])
b = np.array([1,2,3])
print(A, b, sep=2 * '\n')[[42 2 3]
[ 4 5 6]
[ 7 8 9]]
[1 2 3]# solve a system of linear equations
x = np.linalg.solve(A, b)
xarray([2.18366847e-18, 2.31698718e-16, 3.33333333e-01])A.dot(x)array([1., 2., 3.])A = np.random.rand(3,3)
B = np.random.rand(3,3)
evals, evecs = np.linalg.eig(A)
evalsarray([1.68827208+0.j , 0.05361425+0.50545607j,
0.05361425-0.50545607j])evecsarray([[ 0.55636511+0.j , 0.62887518+0.j ,
0.62887518-0.j ],
[ 0.50197522+0.j , -0.4598983 +0.42547761j,
-0.4598983 -0.42547761j],
[ 0.66217727+0.j , -0.2385688 -0.39378077j,
-0.2385688 +0.39378077j]])Decomposes any matrix \(A \in \mathbb R^{m \times n}\) as follows: \[ A = U \times S \times V^\top \] where - \(U\) and \(V\) are orthonormal matrices (meaning that \(U^\top \times U = I\) and \(V^\top \times V = I\)) - \(S\) is a diagonal matrix that contains the singular values in non-increasing order
print(A)
U, S, V = np.linalg.svd(A)[[0.74195289 0.87151047 0.13443881]
[0.1871254 0.32101526 0.87924946]
[0.82629926 0.34492737 0.73253243]]U.dot(np.diag(S)).dot(V)array([[0.74195289, 0.87151047, 0.13443881],
[0.1871254 , 0.32101526, 0.87924946],
[0.82629926, 0.34492737, 0.73253243]])A - U @ np.diag(S) @ Varray([[ 0.00000000e+00, -3.33066907e-16, -3.05311332e-16],
[-1.66533454e-16, -1.66533454e-16, 0.00000000e+00],
[-1.11022302e-16, -4.44089210e-16, 1.11022302e-16]])# U and V are indeed orthonormal
np.set_printoptions(precision=2)
print(U.T.dot(U), V.T.dot(V), sep=2 * '\n')
np.set_printoptions(precision=8)[[ 1.00e+00 -1.24e-16 1.15e-16]
[-1.24e-16 1.00e+00 3.46e-16]
[ 1.15e-16 3.46e-16 1.00e+00]]
[[1.00e+00 3.45e-16 1.52e-16]
[3.45e-16 1.00e+00 1.59e-17]
[1.52e-16 1.59e-17 1.00e+00]]scipy.misc.face()compute_approx(X, r)!pip3 install poochRequirement already satisfied: pooch in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (1.8.2) Requirement already satisfied: platformdirs>=2.5.0 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from pooch) (4.3.6) Requirement already satisfied: packaging>=20.0 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from pooch) (24.2) Requirement already satisfied: requests>=2.19.0 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from pooch) (2.32.3) Requirement already satisfied: charset-normalizer<4,>=2 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from requests>=2.19.0->pooch) (3.4.1) Requirement already satisfied: idna<4,>=2.5 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from requests>=2.19.0->pooch) (3.10) Requirement already satisfied: urllib3<3,>=1.21.1 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from requests>=2.19.0->pooch) (2.3.0) Requirement already satisfied: certifi>=2017.4.17 in /home/boucheron/Documents/IFEBY310/.venv/lib/python3.12/site-packages (from requests>=2.19.0->pooch) (2024.12.14) [notice] A new release of pip is available: 25.0 -> 25.1.1 [notice] To update, run: pip install --upgrade pip
import numpy as np
from scipy.datasets import face
import matplotlib.pyplot as plt
%matplotlib inline
X = face()type(X)numpy.ndarrayplt.imshow(X)
_ = plt.axis('off')
n_rows, n_cols, n_channels = X.shape
X_reshaped = X.reshape(n_rows, n_cols * n_channels)
U, S, V = np.linalg.svd(X_reshaped, full_matrices=False)X_reshaped.shape(768, 3072)X.shape(768, 1024, 3)plt.plot(S**2) ## a kind of screeplot
plt.yscale("log")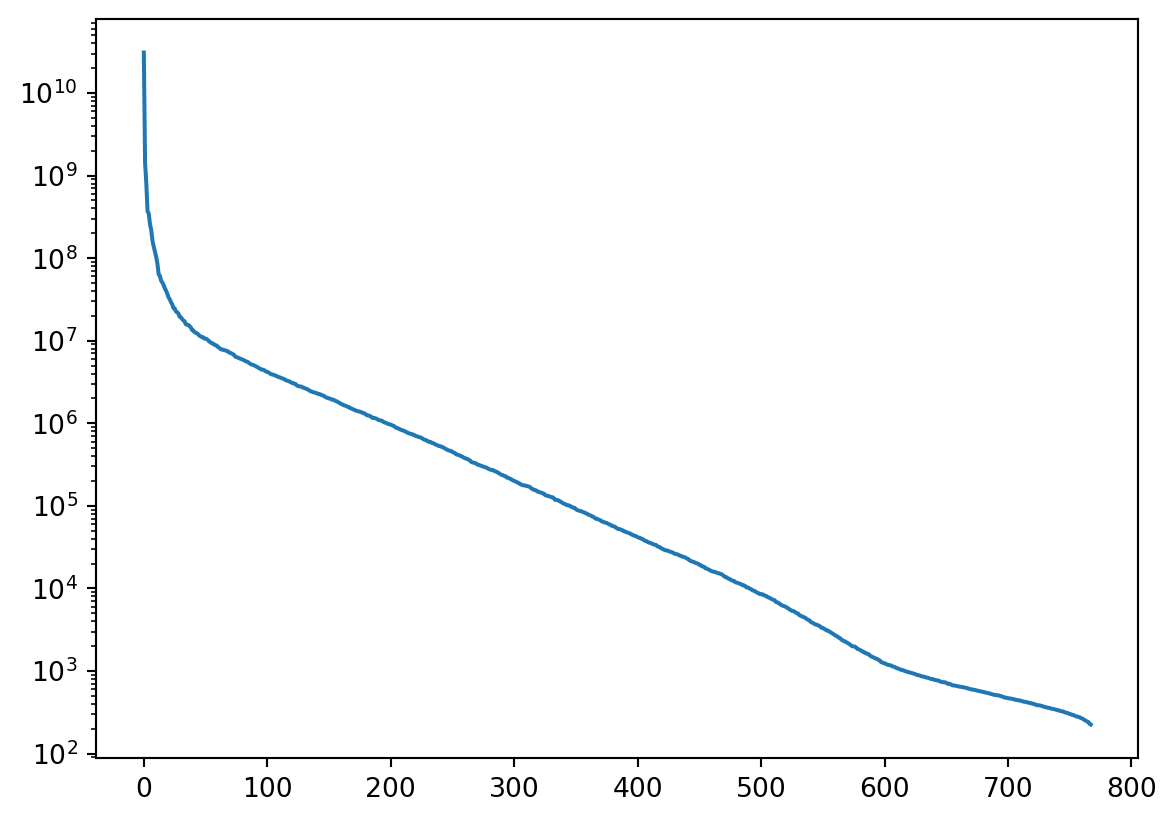
def compute_approx(X: np.ndarray, r: int):
"""Computes the best rank-r approximation of X using SVD.
We expect X to the 3D array corresponding to a color image, that we
reduce to a 2D one to apply SVD (no broadcasting).
Parameters
----------
X : `np.ndarray`, shape=(n_rows, n_cols, 3)
The input 3D ndarray
r : `int`
The desired rank
Return
------
output : `np.ndarray`, shape=(n_rows, n_cols, 3)
The best rank-r approximation of X
"""
n_rows, n_cols, n_channels = X.shape
# Reshape X to a 2D array
X_reshape = X.reshape(n_rows, n_cols * n_channels)
# Compute SVD
U, S, V = np.linalg.svd(X_reshape, full_matrices=False)
# Keep only the top r first singular values
S[r:] = 0
# Compute the approximation
X_reshape_r = U.dot(np.diag(S)).dot(V)
# Put it between 0 and 255 again and cast to integer type
return X_reshape_r.clip(min=0, max=255).astype('int')\
.reshape(n_rows, n_cols, n_channels)ranks = [100, 70, 50, 30, 10, 5]
n_ranks = len(ranks)
for i, r in enumerate(ranks):
X_r = compute_approx(X, r)
# plt.subplot(n_ranks, 1, i + 1)
plt.figure(figsize=(5, 5))
plt.imshow(X_r)
_ = plt.axis('off')
# plt.title(f'Rank {r} approximation of the racoon' % r, fontsize=16)
plt.tight_layout()





In the code above, we recompute the SVD of X for every element in list rank.
In the next chunk, we compute the SVD once, and define a generator to generate the low rank approximations of matrix X. We take advantage of the fact that the SVD defines an orthonormal basis for the space of matrices. In this adapted orthonormal basis the optimal low rank approximations of \(X\) have a sparse expansion.
def gen_rank_k_approx(X):
"""Generator for low rank
approximation of a matrix X using truncated SVD.
Args:
X (numpy.ndarray): a numerical matrix
Yields:
(int,numpy.ndarray): rank k and best rank-k approximation of X using truncated SVD(according to Eckart-Young theorem).
"""
U, S, V = np.linalg.svd(X, full_matrices=False)
r = 0
Y = np.zeros_like(X, dtype='float64')
while (r<len(S)):
Y = Y + S[r] * (U[:,r,np.newaxis] @ V[r,:, np.newaxis].T)
r += 1
yield r, Yg = gen_rank_k_approx(X_reshaped) for i in range(100):
_, Xr = next(g)
if i % 10 ==0:
plt.figure(figsize=(5, 5))
plt.imshow(
Xr
.clip(min=0, max=255)
.astype('int')
.reshape(n_rows, n_cols, n_channels)
)
_ = plt.axis('off')
plt.tight_layout()









Visit https://numpy.org/numpy-tutorials/content/tutorial-svd.html